There’s never a dull moment.
Earlier today, I read a paper from Nature titled “Nascent transcript sequencing visualizes transcription at nucleotide resolution*”. The paper describes a very cool new technique that allows you to see gene transcript sequences as they are being made. The authors demonstrate that this method can be used to sequence unstable, short-lived pre-modification mRNAs; quantify sense and antisense transcript abundance; identify proteins that regulate promoter directionality; and map transcription pause sites.
Cool stuff, right?!
(If you’re not in this field: trust me on this. It’s megasupercool).
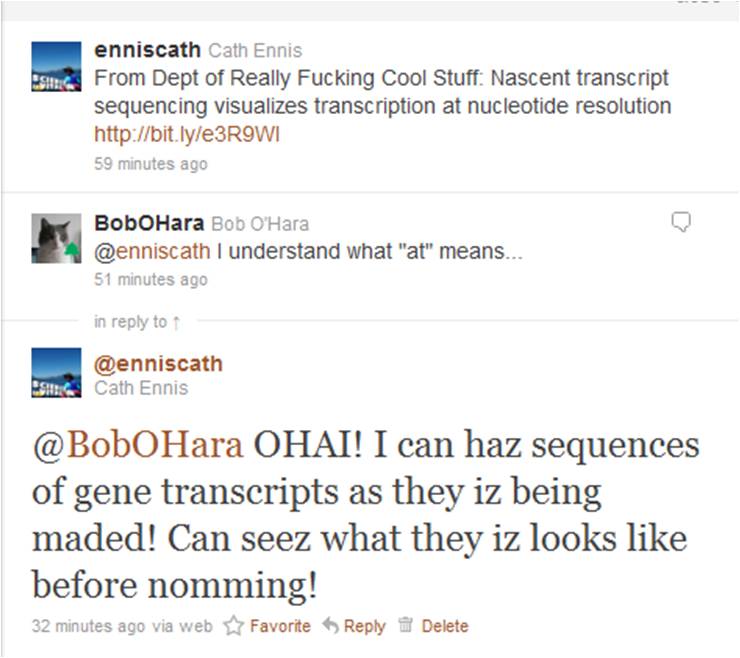
So, this article being in the same issue of Nature as a feature on rapid online post-publication review (“Peer review: Trial by Twitter“), I obviously had to tweet about it. And that’s when the fun began:
In no time at all, Bob maded me a LOLcat, and then he eated tweeted it:
Genius! Ribosome Polymerase Kitteh iz teh LOLz!
The Nature feature on online peer review says:
“Unstructured, unruly and often anonymous, online commenting can be exasperating for biologists used to more conventional means of discussion”.
I can’t possibly think why.
……………………………………….
*Full citation: Churchman LS, Weissman JS. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature 2010: 469:368-373.
I’d also just like to say that Stirling Churchman is an awesome name.


Genius! I love it!
That sums it up so clearly that you should seriously consider adding the Research Blogging icon. 🙂
Heh! I’m not sure it’s quite their cup of tea, but thanks!
SUPER! This paper was totally awesome, and I think the LOLcat is perfect. Too bad they didn’t use that as the “visual abstract”!!
btw, full name is Lee Stirling Churchman. Check out her website: http://www.leestirlingchurchman.com/Info/About.html. There is a PIE tab!!
PIE! PIE AND MUSIC!
ZOMG, NEW SCIENCE CRUSH!
Nicely spotted, Gerty, and thanks for sharing that!
Teh kittehs is teh awesomesauces.
Thanks all! I already have 4 more comments than the Nature article about online commenting!
Maybe if Nature subtly and sneakily combined substance with silly, they’d get more comments. 😉
Cath,
I hadn’t seen the “Trial by Twitter” commentary, so I just finished reading it. I was in shock when I read the following comment:”It makes much more sense in fact to publish everything and filter after the fact,” says Cameron Neylon, a senior scientist at the Science & Technology Facilities Council, a UK funding body.
This would be an absolute disaster!
I agree that peer review is far from perfect–but like democracy, there doesn’t seem to be a better way. I speak from experience in saying that there is NO WAY that it would ever be possible to “filter after the fact”. The reason for this is that research today is so vast, that only researchers really in the field (and particularly those doing the peer review) actually read the fine detail enough to make judgements about what to filter. The rest are only looking for the “take home message”–and if they are going to take home that message when it hasn’t been minimally approved by basic peer review–we will end up setting scientific research back several decades. As it is, even with peer review, bad papers sneak through the cracks and confuse us with their incorrect interpretations or improperly controlled experiments. Who would have time to have to examine every paper published like a peer reviewer in order to build on others results?
This comment really frightened me…
The quote is really a bit out of context in the way its presented in the article but nonetheless I’ll stand by it in that form. The bottom line with peer review in its current form prior to publication is that there really isn’t any good evidence that it achieves anything. What there is is good evidence that it costs a lot of money. If you accepted peer review worked and relied on just the take home measures then you’d be thinking that there are microbes with arsenic in their DNA, that hydrides can oxidize benzaldehyde and that there are known gene associations with longevity. Bottom line if you don’t look critically at any paper in any venue then you’re a poor scientist. When this happens en masse we have a poor science community.
What I find striking is that it is possible to have rapid, cheap, and critical analysis of these papers after they are published. It would also be trivial to capture that information and present it back at the paper. The problem lies in the fact that people aren’t really willing to do this except for really obvious mistakes on really big papers. So to make it work you’d have to have a broader community doing some sort of rating and ranking across the broad range of papers. And we’d have to learn to be polite rather than just sniggering in coffee roms [none of that Henry!].
But the current system is unsustainable. We either need to do less peer review prior to publication, publish a lot less, or capture rating and ranking information after publication. My contention is that if we go the latter route then we can build something that does for the science literature what Google did for the web. But that works precisely because there is a mass of stuff, some good some bad.
Conversely we could just get Henry to rate everything. That clearly works well for me here 🙂
Hello Cameron,
Nice to meet you virtually (always good to meet an A1 bloke!), and apologies if I took your quote out of context. However (just like in the journal reviews, it’s the “however” that comes in for the kill…)–I am certain that you are quite wrong on this point. It would be easier to refer you to the 2 most recent blogs on peer review (see “Confessions” and “Girl, Interrupting”), but I would like to respond nonetheless.
Yes, occasionally (even in high tier journals) peer review can be suboptimal and papers can “slip through the cracks”. If they can be ‘caught’ afterward by Twitter or scientists on other social media–fine. That’s a bonus.
But, peer review DOES work. I have reviewed, perhaps, hundreds of papers, and am an editor on PLoS One–and I can tell you that overall, the system works quite well. As an editor, very frequently there is an excellent correlation between all 3 reviewers, and in cases where there isn’t, as an editor it is often easy to see which reviewer ‘slacked off’ and did not do a thorough job.
In addition, I completely disagree with your comment that most experiments from other papers cannot be repeated. I’ve been in this business for a few years, and I find it exceedingly rare when we cannot reproduce the main thrust of others’ work. In fact, a major part of our research is simply built upon competition with other labs, each using data published to go the next step and beyond.
I can tell you first hand that if all papers were to be published without peer review, it would be impossible for us to know what is correct and what is garbage. Sure, some things pop out as being utter rubbish, but science is so specialized and diversified, that this it is simply not possible for scientists to make judgements in areas that are not their specialty.
Let’s take a potential example. I am a cell biologist who works on endocytic trafficking. I work in close collaboration with a colleague who is a structural biologist and does a lot of NMR studies. When structures of endocytic regulatory proteins (and interpretations of the structures) are published, I rely on them as a resource to build on my own studies. But do I fully understand the fine lines in the methodologies of NMR and crystal data? No, I do not. So if the paper comes out having been peer reviewed in Henry’s favorite weekly, I rely on the integrity of the authors and peer reviewers in order to decide what mutants might be interesting to test in our pathways. If the paper were to come out without being peer reviewed, how would I have any way of knowing whether the data and proposed model are likely to be valid or simply rubbish?
The system you seem to propose would really necessitate scientists to “reinvent the wheel” every time they need to set up an experiment.
I simply do not buy your argument that “peer review does not work”–the bulk of meaningful scientific data that pushes science and technology forward is derived from the peer review system. Sure, it’s not perfect. Sometimes it’s frustrating when papers are not accepted, when there are vindictive reviewers with competing interest. But overall, it’s like democracy–not perfect, but compared to anything else, it’s the best there is!
By the way, I have no hidden agenda or policy. I’m just a scientist doing my research, teaching and mentoring. But I truly do want to convince you to think again about this issue, as I think that it’s really a crucial one for scientists, and probably something that the general public is not fully able to appreciate.
Thanks for your input!
Steve
Steve, just came back here in the process of writing my own (hopefully explicative) blog post but I wanted to pick up two things. First if you actually look at objective evidence (and not anecdote) we really have essentially no evidence that peer review achieves any of things you say it does. It is difficult of course to show an effect but no-one has succeeded yet. That should worry us.
Also key to my argument is not that we throw away peer review but that we apply it more efficiently. Its effectively our most precious resource and what I’m arguing is that rather than use it (fairly ineffectually) to block publication at a very specific (and fairly arbitrary) time point in the research cycle we should publish everything (which basically costs us nothing) but then carry out continuous review over time. Nothing gets the quality stamp until its received some minimum number of reviews, but equally someone can come along three years later and note that it turns out to be wrong. Or someone who’s actually tried the method can comment that it doesn’t work.
My problem is that we’re currently effectively throwing information away when we don’t need to, while not actually doing a good job of utilising the effort that is put into review to get its full value. If you want a simple straw man argument. What would be wrong with publishing everything, then doing the peer review, then re-publishing in “the journal”. Works in both physics and atmospheric chemistry. What I’m absolutely not saying is that we throw peer review totally out of the window.
Cameron Neylon is well known to many of us personally. He is an A1 good bloke. He is however something of an open-science evangelist. I agree with you, Steve. I think open peer review isn’t the way to go. But I would say that, wouldn’t I?
Henry: How does the saying go? “Some of my best friends are open-science evangelists…”
I have no doubt he’s a great bloke–but I can think of nothing worse than having my lab work for months on something that’s supposed to build on another scientist’s work and go a step farther–only to find that the conclusions were wrong, or can’t be repeated or some other issue that crops up because the paper was never peer reviewed. As it is, that still happens occasionally and “takes the piss out of us”. But to set up a system where it is this way intentionally?
Glad you agree–no bias, eh? BTW, my kids keep wondering around singing “vertebrae, oh vertebrae” all day thanks to you…
And sorry Cath– I did move this off the beaten track– a very funny (silly? silly=funny) post. Sorry for the digression, but that really perked up my antennae.
There’s no need to ever apologise to me* for off-topic comments. It’s all part of the joy of blogging!
*some other bloggers may disagree, and may even berate people for a seemingly on-topic comment about the wrong part of their post. I’ve seen it happen!
Buses. Stations. Girrafes on unicycles. You know the drill.
Maybe I should have replied in another order. Steve, if you haven’t had this happen to you, with supposedly well reviewed papers in reputable journals, then you’ve never worked in a lab. I don’t think I’ve ever managed the replicate anything from a paper without having to apply some tweaks.
But worse, I wasted two years of a postdocs life on trying something that a whole bunch of other people had already tried and knew didn’t work. But why go through the grief of peer review to show something doesn’t work? Waste of effort, so I never knew until it was too late. And I have I published it? Or course not, why would I bother?
But if it was simple to publish. And then a few other people just commented on it that they’d also had problems, then we could have saved £150k of taxpayers money and done something useful with it…
Reading “It makes much more sense in fact to publish everything and filter after the fact” just made me panic. Even some people I work with (those with above average understanding of biology and immunity) are still hesitant to have their kids vaccinated – just in case the autism link is real. After all, it was published wasn’t it?
Publish everything and filter later = Scientific Enquirer? No, actually, at least they seek legal advice in advance to minimize the chances of being sued.
(Sorry to hijack the thread, Cath, the tweets and accompanying LOL made me chuckle audibly). I will have to read that article.
Cath
Stirling Churchman gives me an idea for a new game (and Stirling is a second name? wonder what the first one was they were trying to avoid?)
Favourite scientist names game (optional rule: must be in pubmed to qualify?)
My personal favourite: Fabio Pizza.
Now *this* is what the internet is all about!
Ruchi, silly? Sure. Subtle? Who, me? 🙂
Steve, CromerCrox & Mermaid: I agree, mostly. Mermaid’s vaccine story is scary, and I’m gonna have to find out who these people are now!
Antipodean, that’s an awesome name! In my postdoctoral field, one of the big names was Jerzy Jurka. I’m sure at least one of the Js is pronounced as a Y in real life – but not in my head!
Sarah, welcome to the blog! Glad you approve of the silliness; there’s a lot of it about round here.
OMG! For a brief moment, I thought that was a real snake with the LOLcat. Phew!
I’m a fan of interesting author combinations on papers, e.g. Fatt and Katz (1952).
i don’t think it gets much better than Fatt and Katz, actually.
There is a well-known collaboration of scientists that works on skin microflora. Just ask PubMed for ‘Dark and Strange’, or, as it may be, ‘Strange and Dark’.
But my favourite scentist name of all time goes to a very self-effacing chap I once met at Livermore who spends his time peeling electrons off uranium atoms.
Roscoe Marrs.
Fatt and Katz is great!
I once spotted a citation of “Browne and Proud, 2004” that made me smile.
There are probably some excellent ones I’m missing because I don’t speak any Chinese or Japanese.
For Hebrew speakers, the Japanese combo of Nagata and Takata has to stand out. Nagata means “you touched” and Takata means “You… oh, never mind *slinks off embarassed*
Hebrew translations of Japanese? I love the internet!
Wow, this comment thread really is all over the place (more than usual).
Just wanted to chime in with my appreciation and admiration for Bob O’Hara’s ribosome LOL kitty. Oh, and I looked up the first author’s web page thanks to Gerty’s link–I think I have a new science girl crush, too, Cath.
I know, it’s a real mish-mash – isn’t it great?!
I will forever view the ribosome as a snake-eating kitty.
It’s the only way to fly.
It would actually make sense for most cellular mechanisms to be regulated by millions of segments of lolcats. The whole damn business is unruly and makes no fucking sense.
*Like*
+1
Awesome! I hear visual abstracts are the new fashion…
The “filter later” bit on peer review scares me too, though.
Yes, so it would seem. I think LOLcat visual abstracts have about as much chance of catching on as “publish absolutely everything and sort it out later”, though.
I have nothing to add, other than the name of a plant pathologist: “R.C. Shattock”. To be said quickly.
Unfortunate.
There’s an NHL player called Shattenkirk. Wonder if he’s a Star Trek fan?
This is fucking awesome. I’m wondering if I should prepare a LOLcat version of what I do for a living, save it to my iPhone for use when, oh, I don’t know, any single family member or non-science friend ask me what I do for a living.
Do it! (Then blog it!)
I love making up wonderful names for graduate exam questions. Some of my favorites: Dr. Ken E. Tecks, Prof. C.N. Fuge, Dr. D. Tergent, Dr. P.R. Dox, Prof. Dro. S. Phila, Dr. G.W. Knuckular (special fav), Prof. Sal Monella (special fav), Dr. V. Skull (vesicle…)
Needless to say, I get no appreciation for my efforts….
Hi, I’m really confused about this point. The original Wakefield paper was peer reviewed. This is kind of my point. Peer review doesn’t catch this kind of thing, its really really bad at precisely that job. We’d do much better to formalize the process of capturing all the expert opinion after e.g. this paper was published and make it visible on the paper (467 out of 467 experts thought this paper was a pile of tosh). If that paper had been published today you would have had dozen of blog posts within hours pointing out the holes. Isn’t that (at least potentially, assuming a whole bunch of things) a more effective way of identifying the problems that peer review failed to catch?
This is a repeat of my response from above:
Hello Cameron,
Nice to meet you virtually (always good to meet an A1 bloke!), and apologies if I took your quote out of context. However (just like in the journal reviews, it’s the “however” that comes in for the kill…)–I am certain that you are quite wrong on this point. It would be easier to refer you to the 2 most recent blogs on peer review (see “Confessions” and “Girl, Interrupting”), but I would like to respond nonetheless.
Yes, occasionally (even in high tier journals) peer review can be suboptimal and papers can “slip through the cracks”. If they can be ‘caught’ afterward by Twitter or scientists on other social media–fine. That’s a bonus.
But, peer review DOES work. I have reviewed, perhaps, hundreds of papers, and am an editor on PLoS One–and I can tell you that overall, the system works quite well. As an editor, very frequently there is an excellent correlation between all 3 reviewers, and in cases where there isn’t, as an editor it is often easy to see which reviewer ‘slacked off’ and did not do a thorough job.
In addition, I completely disagree with your comment that most experiments from other papers cannot be repeated. I’ve been in this business for a few years, and I find it exceedingly rare when we cannot reproduce the main thrust of others’ work. In fact, a major part of our research is simply built upon competition with other labs, each using data published to go the next step and beyond.
I can tell you first hand that if all papers were to be published without peer review, it would be impossible for us to know what is correct and what is garbage. Sure, some things pop out as being utter rubbish, but science is so specialized and diversified, that this it is simply not possible for scientists to make judgements in areas that are not their specialty.
Let’s take a potential example. I am a cell biologist who works on endocytic trafficking. I work in close collaboration with a colleague who is a structural biologist and does a lot of NMR studies. When structures of endocytic regulatory proteins (and interpretations of the structures) are published, I rely on them as a resource to build on my own studies. But do I fully understand the fine lines in the methodologies of NMR and crystal data? No, I do not. So if the paper comes out having been peer reviewed in Henry’s favorite weekly, I rely on the integrity of the authors and peer reviewers in order to decide what mutants might be interesting to test in our pathways. If the paper were to come out without being peer reviewed, how would I have any way of knowing whether the data and proposed model are likely to be valid or simply rubbish?
The system you seem to propose would really necessitate scientists to “reinvent the wheel” every time they need to set up an experiment.
I simply do not buy your argument that “peer review does not work”–the bulk of meaningful scientific data that pushes science and technology forward is derived from the peer review system. Sure, it’s not perfect. Sometimes it’s frustrating when papers are not accepted, when there are vindictive reviewers with competing interest. But overall, it’s like democracy–not perfect, but compared to anything else, it’s the best there is!
By the way, I have no hidden agenda or policy. I’m just a scientist doing my research, teaching and mentoring. But I truly do want to convince you to think again about this issue, as I think that it’s really a crucial one for scientists, and probably something that the general public is not fully able to appreciate.
Thanks for your input!
Steve
@Cameron – yes, there are egregious examples of peer-reviewed papers that were wrong, but these are newsworthy in part because they are so few – there is no need to use these as a reason for abolishing the whole system. Another point perhaps worth making is that the publishing ecosystem is increasingly diverse. Many journals are peer-reviewed. Some are not. Some are in print, others are online. Some disciplines are happy with preprint severs, others tend not to be. Some journals publish papers for free and charge a subscription, with others it’s pay to play. In such a marketplace, then there is nothing to stop you or anyone else setting up a platform that accepts all papers and relies on peer-review after the fact.
My silliest posts have a tendency to attract many very serious comments. I wonder if I can get a grant to study this phenomenon.
The nature piece complained that people didn’t comment on papers on the journal’s pages. I feel a letter to Nature is in order, to explain what they’re doing wrong. With LOLcat, of course.
Let’s do it! That would be hilarious!
I think it’s interesting that Cameron and Steve are having trouble reproducing each other’s experiences of reproducibiility.
We could probably get a grant to study that, too.
Or a Grant, at the very least.
A comment on the original post – Stirling Churchman is giving a seminar at our institute today! Super excited to see how the original work measures up to the LOLcat graphical abstract 🙂
Oh, cool! Please come back and let me know how it was!
It was a very impressive talk. Anyone who goes from a PhD in single-molecule biophysical stuff to mega-high-throughput sequencing of bits of RNA (while maintaining a sideline in pie experimentation) has got to be pretty special. Lots of clever graphs… From the frequency of detection of each sequence you can infer the direction in which genes are transcribed, and exactly where the machinery pauses at nucleosomes (‘Where does it go? Where does it stop?’)
(Technical note: teh kitteh in above pic actually represents RNA polymerase, not the ribosome….coz LOLcatz iz nothing if not scientifically accurate, even though molecular biologists still mix up transcription and translation occasionally!)
Glad to hear that my new science crush is Teh Awesomez!
Good call on the ribosome. GAAH! I was overcome by enthusiasm and forgot to stop and check for obvious mistakes. Oopsy!